Computation
Computational - Research
Biomedical platforms provide scalable and interoperable infrastructure and secure and compliant services to ingest, process, validate, integrate, curate, store, analyze and share data. These platforms support workflows, data analyses, visualization tools, and access to storage repositories.
Today most large-scale biomedical platforms use secure cloud computing technology for analyzing and storing phenotypic, clinical, and genomic data. Many web-based platforms are available for researchers to access services and tools for biomedical research. The use of containers such as Docker Containers facilitates the integration of bioinformatics applications with various data analysis pipelines. Adopting Common Data Models (CDMs), Common Data Elements (CDEs), and Ontologies enables reuse.
Common Data Models (CDMs) are used to standardize data collection, facilitating the aggregation and sharing of data. There are several CDMs available for specific uses, and examples include the National Patient-Centered Clinical Research Network (PCORnet) and the Observational Medical Outcomes Partnership (OMOP). An evaluation of CDM use for longitudinal Electronic Health Record (EHR) based studies showed that the OMOP CDM best met the criteria for supporting data sharing. The "All-of-Us" (AOU) program is standardizing EHR data by using the OMOP CDM. Methods to harmonize data from the i2b2/ACT system to the OMOP CDMare also available.
Common Data Elements (CDEs) are used in clinical research studies and represent a combination of precisely defined questions (variables) associated with a specified value, i.e., the Common Data Element. The use of CDEs benefits data aggregation, meta-analyses, and cross-study comparisons.
Data at scale requires the development of strategies that can efficiently leverage public cloud computing resources. Research is advanced through the use of research community developed standards for data collection. It is also advanced by developing machine learning methods that support data integration from multiple disease areas.
The FAIR principles were promoted to make data findable, accessible, interoperable, and reusable. The TRUST principles encourage the adoption of Transparency, Responsibility, Usability, Sustainability, and Technology. These principles are complementary to the FAIR principles and aim to promote TRUST for data repositories.
Trustworthiness is demonstrated through evidence, which depends on transparency, and thus repositories must provide transparent, honest, and verifiable proof of their practice. In this way, stakeholders can be confident that repositorieswill deliver dataintegrity, authenticity, accuracy, reliability, and accessibility over extended time frames. Trustworthiness must include regular audits and certifications.
Cloud-based Platforms and Ecosystems
In the past, data processing in biosciences required the support of many styles of computation. Datacenter computing supported batch and grid workloads. Platforms were configured with various storage deployments (DAS, NAS, Disk Storage Arrays, etc.). These server platforms typically consisted of large symmetrical multiprocessing (SMP) hosts and relational databases. This scale-up technology was "best practice" 15-20 years ago. While this form of server technology was well suited to structured biomedical data sets, it was ill-suited to the genomic data explosion and today's heterogeneous data types. It was burdened with older commercial database architectures. These deployments have been cumbersome at best, and these server platforms require a great deal of support. This is especially true when the data is organized as a data warehouse and requires some script-based extract, transfer, and load (ETL) process to collocate other datasets from other environments. This is no longer practical for dynamic large-scale data environments where data is simply too big to move.
Bioscience researchers recognized the need to support ever-growing petabyte-scale data sets. They sought to take advantage of improvements in processing power and lower compute costs. They realized that "one size does not fit all." They sought platforms to support international research efforts and international collaboration. These researchers saw advantages in platforms consisting of high-performance data centers and commercial clouds, seamlessly integrated and performing as single entities.
Consistent with this desire, the next-generation platform must support data-intensive computation. We can envision biomedical clouds where terabytes of new data would be added each day and processed each night, and available for search the following day. It must provide the needed computational capability to perform any necessary integer or floating-point calculation, data movement, encryption of data, and data analytics.
This computational capability must exist regardless of the computational framework available for a specific style of data analysis. Such terms have defined these frameworks and styles of computing as SQL, Non-SQL, Graph Databases, Time-series Databases, shared-nothing architectures, 3-tier computing, Data Warehouses, VLDB, in-memory DB, Field Programmable Gate Arrays, Array Databases (multi-dimensional array models), GRID computing, Message Queues, Map Reduce (Hadoop), Apache Spark, GPUs, Distributed Computing, Clusters, Parallel Processing, Federated Data, web-services, SMP, PaaS, SaaS, IaaS, Software-defined, storage, networking or datacenters, Supercomputing, etc.
The platform must match computation to the researchers' needs, given the data. Cost-effectiveness is important. Some data may best use data center computation (transactions), while other computation forms may be most cost-effective in the cloud. Based on the workload, hybrid clouds may deliver the most cost-effective computation.
Performance metrics used for servers and specialized processors (GPUs, etc.) are no longer relevant in heterogeneous computation environments where a computational pipeline may call upon several different processing types to complete their tasks.

Interoperability and Scalability
Interoperability is the ability of data from non-cooperating resources to integrate and work together with minimal effort. To the greatest extent possible, the next-generation platform must allow interaction with public and private data sets and data sources, medical and bioscience databases, health administration data, research data, and publication data. Examples of limited interoperable databases might include NCBI's Database of Genotypes and Phenotypes (dbGaP) - https://www.ncbi.nlm.nih.gov/gap
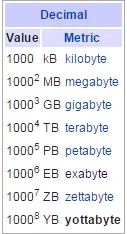
Next-generation interoperability must include public clouds and must be able to move between cloud service providers (CSPs). It must allow for complex analysis that spans multiple cloud services, both public and private. This interoperability must be global. The platform must support tools that will enable this interoperability, including API and Restful Services and notebooks such as Jupyter notebooks. The platform must incorporate reference data sets and reference knowledge bases – such as the latest medical research journals, publications, pub med, etc. Interoperability must be within the constraints of global compliance regulations, codes, laws, and mandates, which differ from country to country and even within countries. The platform must be able to interoperate with many vital repositories of scientific data. Without the ability to search across all of these databases simultaneously, discovery will be constrained.
Each data repository is bound by existing policies, practices, ethical considerations, and legal, contractual obligations in the territory in which it resides. While it is theoretically possible to make these practices consistent between a group of databases, that task is daunting. Similar issues would involve having a single database assume control of all data in separate data infrastructures. Data federation can sometimes overcome some of these obstacles by allowing the data to remain in place, controlled independently by each organization, while at the same time providing significant scientific and operational conveniences for the user. When connecting repositories, common data definitions and standard patient identifiers are prerequisites. Data may need to be operated on and aggregated to some intermediate level and combined with other aggregated data, and analyzed again. This interoperability extends to workflow creation across data sets and clouds. The cost of removing data from commercial clouds is a barrier to this interoperability.
In clinical healthcare, interoperability is the ability of different information technology systems and software applications to communicate, exchange data, and use the information that has been exchanged. Data exchange schema and standards should permit data to be shared across clinicians, lab, hospital, pharmacy, and patient regardless of the application or application vendor. Interoperability means a health information systems' ability to work together within and across organizational boundaries to advance healthcare delivery for individuals and communities.
Seamless interoperability means that the platform you are interoperating with appears as if it is part of your existing system and behaves no different with regard to application data access and speed of computation. The system you interoperate with and its data must appear the same as if that data were on your own system. Any seams must be invisible.
Effective data stewardship and effective data management mean using community-agreed data formats, metadata standards, tools, and services that can improve data integration capabilities.
New insights in disease area research will require platforms to support integrating multiple data types - genomics, proteomics, imaging, phenotypic and clinical data for research projects. New modalities will also require addressing the challenge of determining commonalities across various disease area research.
The explosion of biological data is overwhelming centralized systems' ability to handle them in a capacity or computational sense. The platform must scale computationally. It must scale total platform memory and combined disk storage, and combined network bandwidth. More than this, the systems and application software must scale in the number of concurrently executing processes/threads. The platform must also scale globally. Available analytics and system management, and editing tools must also scale. The platform must scale without incurring latency costs for searching larger and largerdata sets. It should support parallel file systems and other parallel constructs. We don't want to be limited by the application architecture of the platform.
Scalability also means adding racks of storage, processor memory, and switches in a way that allows you to perform the exact computation in the same amount of time over more data.
Research Workspaces
A team of researchers' effectiveness depends heavily on their ability to collaborate while leveraging shared applications and tools in a secure and protected environment. Towards this end, researcher workspaces should be extensible, customizable, and easy to use. An environment would enable:
- Researchers'ability to interact with their environment as if it were a virtual cluster. Users should be able to scale up and scale down their virtual clusters at runtime with just a single operation in the browsers. They should also be able to install any software needed as if they were operating a real cluster.
- Researchers who have different requirements in terms of software packages and configurations should be able to pre-define their own Workspace according to their demands without knowing the details of the underlying infrastructure and automatically provide a cloud workspace based on their requirements.
- Container-based virtualization with advantages like building workspaces in seconds, high utilization of resources, running compute over thousands of containers on a single physical host, etc. With virtualization technology, the workspaces of different users must work together in a public or private cloud. The performance of computing, memory throughput, disk I/O, and network transmission must be guaranteed.
- An extensible plugin architecture that allows individuals and teams to easily add their own data types, algorithms, and GUI components into the workspace framework to use and share with others. It would enable access to popular brain science pipelines and workflows and research libraries and tools (Jupyter notebooks) and to languages such as Javascript, Python, R, Rstudio, and MATLAB. It must provide interoperability between a wide range of software systems and components.
- High levels of operation, component, workflow, and plugin reuse. This reduces the development time and cost of future development. Once within the workspace framework, a software element can be included, called, or connected to another component, with the framework imposing no limits on reuse. Nice to have a GUI too.
- A flexible and powerful execution system enabling continuous, inline interaction with data inputs and outputs at any position in the workflow.
- Distributed executions of workflows. Other execution/deployment models such as batch runs from a command line or embedding workflows (as shared library functions) in other software.
- The execution system must select the most effective computation for the workflow and data, including ingest.
- Support for provenance with the ability to connect to/invoke user-specified provenance standards and implementations. This allows meta-data and executable information to be recorded along with simulation results to provide demonstrable traceability.
- Database support with the ability to connect via generic adapter to most database engines. Both relational databases (e.g., SQL) and non-relational databases (e.g., MongoDB) must be supported.
- Wizards for creating template or prototype code in order to reduce the complexity and software engineering skill level required to develop workflows and applications.
- Cross-platform portability of workspace code, workflows, pipelines, plugins, and Workspace-related data.