Data

Data Types
Biomedical research data, especially focused on platforms for brain science, is rich and varied. Organizing and harmonizing across heterogeneous data is extremely challenging and costly. For brain research, this data may include:
(1) Preclinical Data - Animal studies
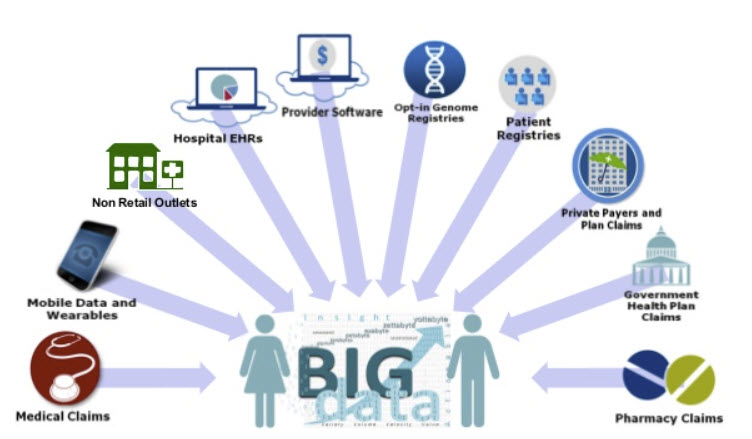
(2) Clinical Data - Clinical data is either collected during ongoing patient care or as part of a formal clinical trial program. Electronic Health Records, Administrative Health Care Compliance data, Insurance Claims information, Patient and Disease Registries, National Health Surveys, Centers for Medicare & Medicaid Services (CMS) data, Bureau of Labor Statistics (BLS) data, and the Census Bureau data are just some of the data sources.
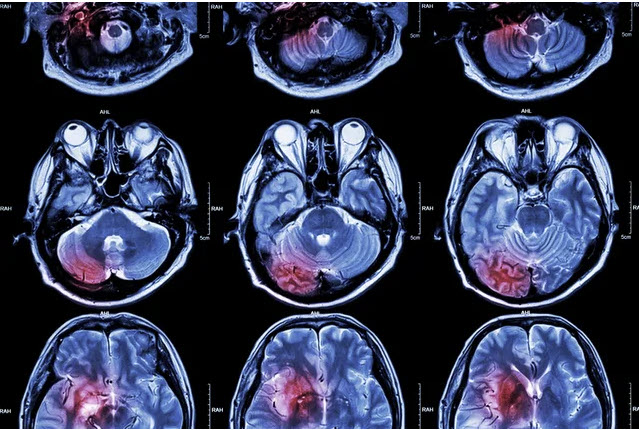
(3) Medical and Biological Imaging Data – Next-generation Brain Science platforms will also support Imaging Data. Biological imaging incorporates radiology which uses the imaging technologies of X-ray radiography, endoscopy, elastography, tactile imaging, thermography, medical photography, and nuclear medicine functional imaging techniques such as positron emission tomography (PET) and Single-photon emission computed tomography (SPECT). An MRI generates a magnetic field around a patient. PET scans use radiopharmaceuticals to create images of the organ's active blood flow and physiologic activity of organs being targeted. As the multimodal and ever higher resolution digital-imaging is embraced across brain science, there will continue to be a swift transition from terabytes to petabytes of data and the need for future cloud tools to manage data much more intelligently.
Some Magnetic Resonance Imaging (MRI) implementations
- fMRI = functional MRI (fMRI)
- rs-fMRI = resting-state functional MRI
- mpMRI = multipoaramtic MRI
- DWI-MRI = diffusion-weighted imaging MRI
- DCE-MRI = dynamic contrast enhancement MRI imaging
- DTI-MRI = diffusion tensor MRI imaging
- DSI-MRI = diffusion spectrum MRI imaging
- rDSI-MRI = radial diffusion spectrum MRI
- MRI-S = spectroscopy
- T1-MRI
- T2-MRI
- structural MRI (T1, T2, PD),
(4) Electrophysiological Data - This includes electroencephalography (EEG), magnetoencephalography (MEG), electrocardiography (ECG), and others. Despite the limited spatial resolution, EEG continues to be a valuable tool for research and diagnosis. It is one of the few mobile techniques available and offers millisecond-range temporal resolution, which is not possible with CT, PET, or MRI. EEG with fMRI is potentially powerful because the two have complementary strengths - EEG has a high temporal resolution and fMRI high spatial resolution.
(5) Genomic and other *omic data
(6) Sensor Data - The platform will also be required to support sensor data (wearables & implantables). Sensor data will play a vital role in baselining conditions before and after treatments and alerting to changes in conditions. Sensor-based data speaks to data volumes that are ingested in real-time.
(7) Social Media and Crowdsourced Data.
(8) Hybrid and Other Modality Data.

Clinical Data Models
There is a great benefit to providing a single common data model for sharing clinical care and observational research information. Examples of existing common data models (CDMs) reflecting diverse data sources and millions of lines of code include:
- National Patient-Centered Clinical Research Network (PCORnet)
- The Sentinel Common Data Model (SCDM)
- Observational Health Data Sciences and Informatics (OHDSI)
- Informatics for Integrating Biology and the Bedside (i2b2)

This long-sought-after common data model for clinical data would have to support the merged common data models of i2b2 Shrine, Sentinel, PCORnet, and OHDSI's OMOP. It would also have to be cognizant of standards development organizations (SDOs) and consortia such as Clinical Data Interchange Standards Consortium (CDISC) and Health Level Seven (HL7).
A common data model would allow clinicians and researchers to pull data from multiple sources and compile it in the same structure without degradation of the information. This endeavor has global implications with the potential to permit the clinical community to define the elements they need, package, and share them in a single consistent structure. This common data model would set the foundation for a mission of global, open-science research and will accelerate the development of effective and safe treatments for disease. It would also free up all the energy and cost used for data harmonization for use elsewhere.
Fast Healthcare Interoperability Resources (FHIR) is a standard describing data formats and elements (known as "resources") and an API for exchanging electronic health records (EHRs). The standard was created by Health Level Seven International (HL7) healthcare standards organization.
FHIR uses a web-based suite of API technology, including an HTTP-based RESTful protocol and a choice of JSON, XML, or RDF for data representation. One of its goals is to facilitate interoperability between legacy health care systems, to make it easy to provide health care information to health care providers and individuals on a wide variety of devices from computers to tablets to cell phones and to allow third-party application developers to provide medical applications which can be easily integrated into existing systems. FHIR provides an alternative to document-centric approaches by directly exposing discrete data elements as services. For example, basic healthcare elements like patients, admissions, diagnostic reports, and medications can be retrieved and manipulated via their resource URLs.
There have been many efforts to bridge and map between specific data models as point solutions.
- Common Data Models (PCORnet CDM, i2b2, OMOP, and Sentinel) to FHIR (FDA/NCATS/CDC/NCI)
- OMOP to FHIR (Mayo)
- Camp FHIR / i2b2 ACT (UNC)
HL7 International and OHDSI announced a collaboration to address the sharing and tracking of data in the healthcare and research industries by creating a single common data model. The organizations will integrate HL7 Fast Healthcare Interoperability Resources (FHIR®) and OHDSI's Observational Medical Outcomes Partnership (OMOP) common data model to achieve this goal. The goal of this project is to facilitate the use of Real-World Data (RWD) sources (e.g., claims, Electronic Health Records (EHRs), registries, electronic Patient Reported Outcomes (ePRO) to support evidence generation for regulatory and clinical decision making.
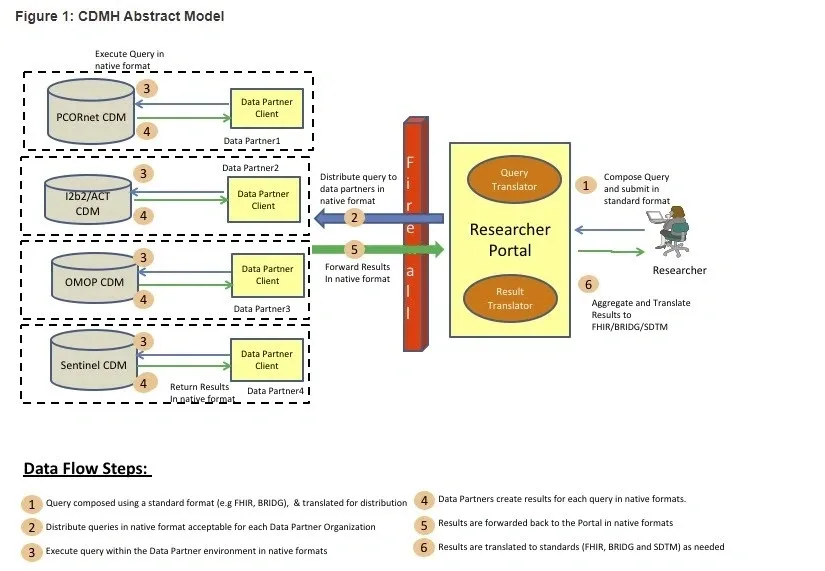
Bridging Common Data Models
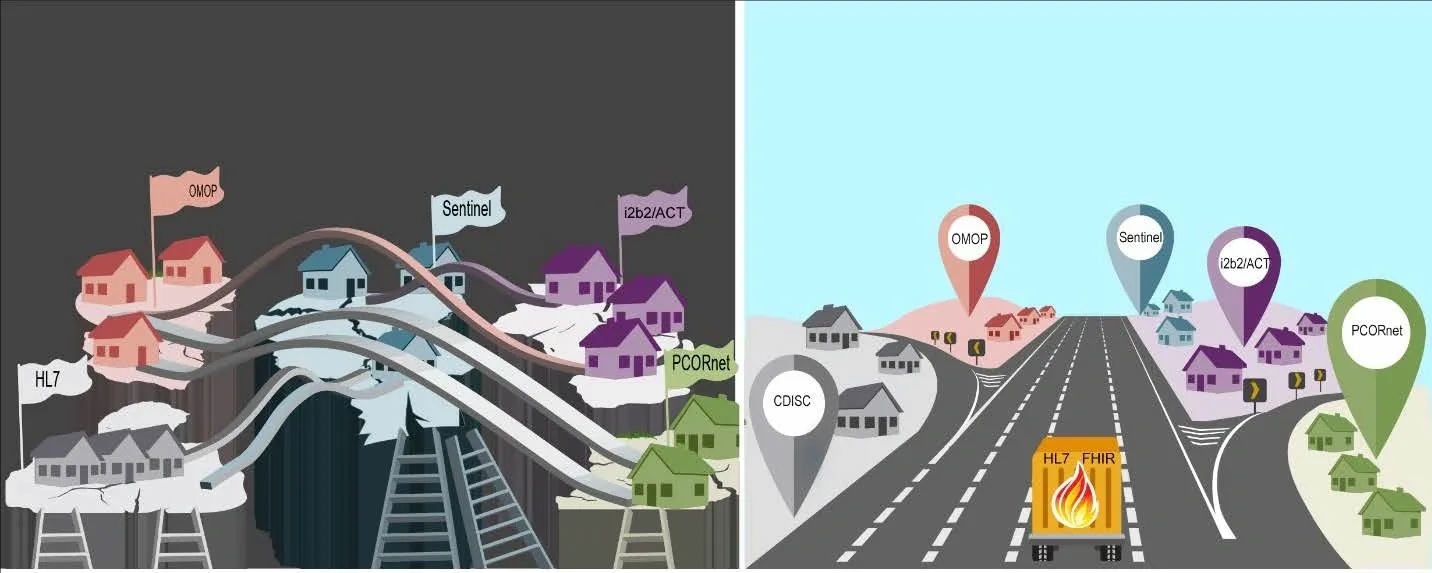
Current and envisioned Common Data Architecture between CDMs
Heterogeneous Data
The next-generation platform will have to curate and compute across a myriad of data types differing in syntax, semantics, file types, representations, and classifications. A clinical study might have targeted tests that might include many data types. For example:
- Targeted Clinical Blood Data
- High Definition Voice Data
- Eye Tracking Data
- Data derived from transcranial magnetic stimulation with electroencephalography, script-driven imagery & psychophysiological reactivity.
- Neurological Optical Coherence Tomography (of the retina) Data.
- Neuro-Imaging Data (fMRI, DSI, rDSI, etc.) including raw DICOM Data.
- Cognitive Test Data.
- Baseline Clinical Interview Data & Self Reporting Data
- Whole sequence Genomic Data

This future platform must be able to concurrently compute (locate, access and process) across these heterogeneous data types and many other categories of data types, including Clinical, EHR, Sensor, Real World, and Patient/Disease repositories.